Evaluation of MALDI-TOF MS for Prediction of Antimicrobial Resistance in Mastitis Associated Bacterial Species
 Reproduced from Ledeboer and Hodinka, 2011
Reproduced from Ledeboer and Hodinka, 2011
Bovine mastitis is the most common, most frequently treated, and most costly infectious disease of dairy cattle. Several infectious agents are associated with clinical and subclinical mastitis in dairy cattle with Staphylococcus aureus being the predominate major pathogen isolated in Canada. Antimicrobial therapy is widely used to treat and prevent mastitis infections in dairy cattle and can decrease the prevalence and duration of mastitis infections. Despite appropriate antimicrobial treatment, bacteriological cure rates seldom exceed 50% for S. aureus, and antimicrobial resistance (AMR) is considered to be one of the reasons for bovine mastitis treatment failures.
β-lactam antimicrobials, including penicillin, cloxacillin, cephapirin, and ceftiofur, are widely used in mastitis treatment protocols. Unfortunately, for most bovine S. aureus mastitis isolates, AMR is due to β-lactamase production. Previous reports on AMR in bovine mastitis associated bacteria indicated that resistance to various antimicrobials is commonly seen in S. aureus mastitis isolates. Cure rate for S. aureus mastitis cases are lower when isolates are penicillin resistant when either β-lactam or non-β-lactam antimicrobials are used. Previous CBMQRN researchers found the prevalence of AMR in S. aureus was 20.4%, with 8.8% of the isolates resistant to penicillin. AMR extends beyond S. aureus, with CBMQRN studies reporting AMR in 25% and 11%, respectively for S. uberis and S. dysgalactiae isolates collected from farms in the Maritime Provinces of Canada.
Traditional methods to detect resistance against antimicrobial drugs are divided into phenotypic and genotypic methods. The phenotypic methods, including the disk diffusion or broth microdilution tests, require overnight incubation, which significantly delays results. Genotypic methods, such as using PCR to detect specific AMR genes, are potentially more rapid but expensive, requiring specialized equipment and laboratory techniques that many diagnostic laboratories do not have access to. Further, molecular tests will only detect the AMR genes for which the assay was designed, thus potentially missing other relevant resistance genes. Thus, there is an urgent need for novel, less expensive methods for rapid detection of AMR. Rapid detection of β-lactamases is important since β-lactam antimicrobials (penicillin, cloxacillin and cephalosporins (cephapirin and ceftiofur)) are the most common antimicrobials used to treat bovine mastitis.
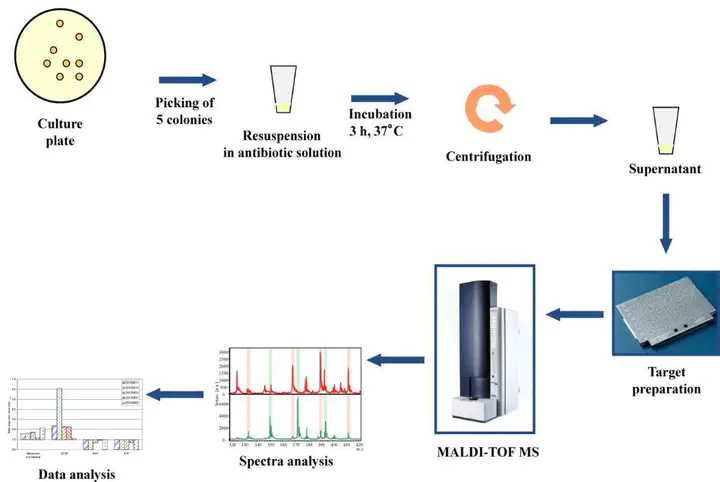
Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) is a rapid, robust, and cost-effective technology that has revolutionized routine microbial diagnostics over the past half-decade. This technology relies on the generation of unique protein profiles (mass spectra) captured from small amounts of bacterial colony material followed by software-assisted searching of a database containing known microbial spectra. MALDI-TOF MS has been widely demonstrated as an accurate method of microbial identification. More recently several studies have focused on other applications of mass spectrometry, such as the detection of AMR in bacteria. A number of approaches have been studied for this purpose in human medicine, including comparison of mass spectra from resistant and susceptible strains, direct detection of the resistance enzyme mass peak and the examination of assay products for detection of antimicrobial degradation. For the latter, a MALDI-TOF MS antimicrobial hydrolysis assay has recently been developed to detect β-lactamase activity by identifying hydrolysis products of β-lactam antibiotics.
We hypothesized that, using well-defined susceptible and resistant S. aureus bovine mastitis isolates from previous research, we can develop a MALDI-TOF β-lactamase assay for rapid, cost effective and accurate determination of β-lactam resistant S. aureus. Following the development of the assay the methodology could be extended to determine β-lactam resistant in other mastitis associated bacteria (e.g. S. uberis, S. dysgalactiae, E.coli and Klebsiella sp.).